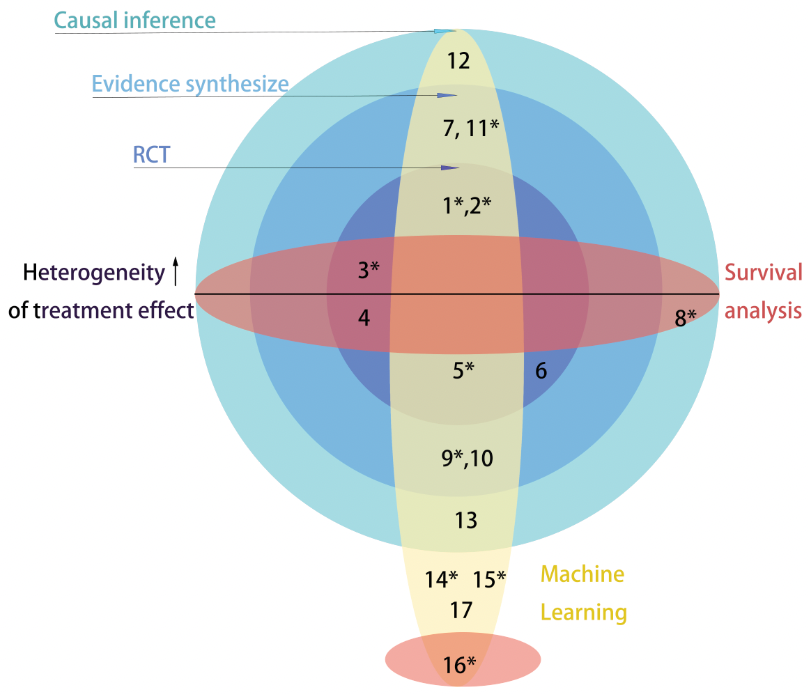
Research distribution, each number represents a published or working paper (see below for the references).
* indicates that I am the first author.
Last updated: 2023.12
My methodological research areas include the intersection of
(1) Causal inference:
(a) Transportability/Meta-analysis: extending inferences from one/more populations to a target population.
(b) Heterogeneity of treatment effects.
(2) Survival analysis: (time-dependent) Cox models and accelerated failure time models with censored outcomes.
(3) Machine learning: Targeted learning and double machine learning, convergence rate characterizing, variable/model selection, and incorporating a priori knowledge into prediction.
Theoretically, I am specialized in non/semi-parametric theory, stochastic process, and optimization.
I have applied the methods and theories into cardiovascular diseases, mental illness, infectious diseases, oncology, and HIV.
* indicates that I am the first author.
Last updated: 2023.12
My methodological research areas include the intersection of
(1) Causal inference:
(a) Transportability/Meta-analysis: extending inferences from one/more populations to a target population.
(b) Heterogeneity of treatment effects.
(2) Survival analysis: (time-dependent) Cox models and accelerated failure time models with censored outcomes.
(3) Machine learning: Targeted learning and double machine learning, convergence rate characterizing, variable/model selection, and incorporating a priori knowledge into prediction.
Theoretically, I am specialized in non/semi-parametric theory, stochastic process, and optimization.
I have applied the methods and theories into cardiovascular diseases, mental illness, infectious diseases, oncology, and HIV.
Grant:
Machine learning for prediction of clinical outcome and estimation of causal effect of direct oral anticoagulants among patients with atrial fibrillation, Fonds de Recherche du Québec Santé (FRQS 272161) doctorate training grant, 70,000 CAD, Role: PI, 2019-2022.
Grant submission:
Lung ultrasound use in the evaluation of the response to fluid management in evolving bronchopulmonary dysplasia: A prospective study, Fonds de Recherche du Québec Santé (FRQS), conditionally approved (Protocol 2023-3517), Role: Collaborator.
Heterogenous Effect Analyses for caRdiovascular Treatments on Survival (HEARTS), NIH K99/R00 Pathway to Independence Award, Role: PI, 2024 Feb. submitted.
Machine learning for prediction of clinical outcome and estimation of causal effect of direct oral anticoagulants among patients with atrial fibrillation, Fonds de Recherche du Québec Santé (FRQS 272161) doctorate training grant, 70,000 CAD, Role: PI, 2019-2022.
Grant submission:
Lung ultrasound use in the evaluation of the response to fluid management in evolving bronchopulmonary dysplasia: A prospective study, Fonds de Recherche du Québec Santé (FRQS), conditionally approved (Protocol 2023-3517), Role: Collaborator.
Heterogenous Effect Analyses for caRdiovascular Treatments on Survival (HEARTS), NIH K99/R00 Pathway to Independence Award, Role: PI, 2024 Feb. submitted.
(10 published)
*Corresponding author; 'co-first author
1. G. Wang*, A. Levis, J. Steingrimsson, IJ. Dahabreh. Efficient estimation of subgroup treatment effects using multi-source data, under review, Statistics in Medicine, arXiv
2. G. Wang'*, S. McGrath', Y. Lian', IJ. Dahabreh, CausalMetaR: An R package for performing causally interpretable meta-analyses, under review, Research Synthesis Methods, arXiv, R package
3. G. Wang, MP. Costello*, H. Pang, J. Zhu, HJ. Helms, I. Reyes-Rivera, RW. Platt, M. Pang, A. Koukounari. Evaluating hybrid controls methodology in early-phase oncology trials: a simulation study based on the MORPHEUS-UC trial, Pharmaceutical Statistics, (2024); 23(1): 31-45. doi:10.1002/pst.2336 Wiley
4. H. Bian*, M. Pang, G. Wang, Z. Lu, Non-collapsibility and Built-in Selection Bias of Hazard Ratio in Randomized Controlled Trials, in revision, BMC Medical Research Methodology, arXiv
5. G. Wang*, A. Levis, J. Steingrimsson, IJ. Dahabreh. Causal inference under transportability assumptions for conditional relative effect measures, under review, Journal of American Statistical Association, arXiv
6. L. Ung*, G. Wang, S. Haneuse, M. Hernán, IJ. Dahabreh. Combining an experimental study with external data: study designs and identification strategies, under review, American Journal of Epidemiology, arXiv
7. Y. Liu, ME. Schnitzer, G. Wang, E. Kennedy, P. Viiklepp, MH. Vargas, G. Sotgiu, D. Menzies, and A. Benedetti*. Modeling Treatment Effect Modification in Multidrug-Resistant Tuberculosis in an Individual Patient Data Meta-Analysis, Statistical Methods in Medical Research (2021) 10.1177/09622802211046383. SAGE
8. G. Wang'*, S. Liu', S. Yang. Continuous-time Structural Failure Time Models with intermittent treatment. arXiv
9. G. Wang, ME. Schnitzer, D. Menzies, P. Viiklepp, TH. Holtz, and A. Benedetti. Estimating treatment importance in multiple drug resistant tuberculosis using Targeted Learning: an observational individual patient data Network Meta-Analysis, Biometrics (2020) 76(3):1007-1016. Wiley
10. AA. Siddique, ME. Schnitzer*, A. Bahamyirou, G. Wang, A. Benedetti et al. Causal Inference for polypharmacy: Propensity score estimation with multiple concurrent medications, Statistical Methods in Medical Research, (2019) 28(12):3534-3549. SAGE
11. G. Wang*. Review 1: ''Antibiotic Prescribing in Remote Versus Face-to-Face Consultations for Acute Respiratory Infections in English Primary Care: An Observational Study Using TMLE.'' Rapid Reviews Infectious Diseases. (2023) RR
12. A. Jaman, G. Wang, A, Ertefaie, M. Bally, R. Lévesque, RW. Platt, and ME. Schnitzer*. Penalized G-estimation for effect modifier selection in the structural nested mean models for repeated outcomes, in revision, Biometrics, arXiv
13. ME. Schnitzer*, A. Ertefaie, D. Talbot, G. Wang, D. Berger, J. O'Loughlin, M.P. Sylvestre. Longitudinal outcome-adaptive and marginal fused Lasso for confounder selection and model pooling with time-varying treatments
14. G. Wang*, ME. Schnitzer, T. Chen, R. Wang, RW. Platt. A general framework for formulating structured variable selection, Transactions of Machine Learning Research, ISSN: 2835-8856 (2024); OpenReview
15. G. Wang*, ME. Schnitzer, RW. Platt, R. Wang, M. Doris, S. Perreault, Integrating complex selection rules into the latent overlapping group Lasso for constructing coherent prediction models, under review, Statistical in Medicine, arXiv
16. G. Wang*', Y. Lian', A. Yang, RW. Platt, R. Wang, M. Dorias, S. Perreault, M. Schnitzer. Structured learning in Cox models with time-dependent covariates, Statistics in Medicine, 2024; 1-20. doi: 10.1002/sim.10116, Wiley, R package
17. A. Bouchard, F. Bourdeau, J. Roger, VT. Taillefer, N. Sheehan, ME. Schnitzer, G. Wang, IJ. Jean Batiste, and R. Therrien*. Predictive Factors of Detectable Viral Load in HIV Infected Patients, AIDS Research and Human Retroviruses, (2022) 38(7):552-560, Libertpub
18. X. Di, Y. Chi, L. Xiang, G. Wang, B. Liao*.Association between Sitting Time and Urinary Incontinence in the US Population: data from the National Health and Nutrition Examination Survey (NHANES) 2007 to 2018, Heliyon (2024) 10(6):E27764, Elsevier
19. G. Wang, TE. Liao, D. Furfaro, LA. Celi*, KS. Ma*, Extending inference from randomized clinical trials to target populations: a scoping review of transportability methods, arXiv, under review, Journal of the American Medical Informatics Association
20. G. Wang, PJ. Heagerty, IJ. Dahabreh*, Effect score analyses in randomized clinical trials, Journal of American Medical Association. 2024;331(14):1225–1226. doi:10.1001/jama.2024.3376, JAMA
21. G. Wang'*, R. Karlsson'*, J. Krijthe, IJ. Dahabreh, Robust integration of external control data in randomized trials, under review, Biometrics , arXiv
*Corresponding author; 'co-first author
1. G. Wang*, A. Levis, J. Steingrimsson, IJ. Dahabreh. Efficient estimation of subgroup treatment effects using multi-source data, under review, Statistics in Medicine, arXiv
2. G. Wang'*, S. McGrath', Y. Lian', IJ. Dahabreh, CausalMetaR: An R package for performing causally interpretable meta-analyses, under review, Research Synthesis Methods, arXiv, R package
3. G. Wang, MP. Costello*, H. Pang, J. Zhu, HJ. Helms, I. Reyes-Rivera, RW. Platt, M. Pang, A. Koukounari. Evaluating hybrid controls methodology in early-phase oncology trials: a simulation study based on the MORPHEUS-UC trial, Pharmaceutical Statistics, (2024); 23(1): 31-45. doi:10.1002/pst.2336 Wiley
4. H. Bian*, M. Pang, G. Wang, Z. Lu, Non-collapsibility and Built-in Selection Bias of Hazard Ratio in Randomized Controlled Trials, in revision, BMC Medical Research Methodology, arXiv
5. G. Wang*, A. Levis, J. Steingrimsson, IJ. Dahabreh. Causal inference under transportability assumptions for conditional relative effect measures, under review, Journal of American Statistical Association, arXiv
6. L. Ung*, G. Wang, S. Haneuse, M. Hernán, IJ. Dahabreh. Combining an experimental study with external data: study designs and identification strategies, under review, American Journal of Epidemiology, arXiv
7. Y. Liu, ME. Schnitzer, G. Wang, E. Kennedy, P. Viiklepp, MH. Vargas, G. Sotgiu, D. Menzies, and A. Benedetti*. Modeling Treatment Effect Modification in Multidrug-Resistant Tuberculosis in an Individual Patient Data Meta-Analysis, Statistical Methods in Medical Research (2021) 10.1177/09622802211046383. SAGE
8. G. Wang'*, S. Liu', S. Yang. Continuous-time Structural Failure Time Models with intermittent treatment. arXiv
9. G. Wang, ME. Schnitzer, D. Menzies, P. Viiklepp, TH. Holtz, and A. Benedetti. Estimating treatment importance in multiple drug resistant tuberculosis using Targeted Learning: an observational individual patient data Network Meta-Analysis, Biometrics (2020) 76(3):1007-1016. Wiley
10. AA. Siddique, ME. Schnitzer*, A. Bahamyirou, G. Wang, A. Benedetti et al. Causal Inference for polypharmacy: Propensity score estimation with multiple concurrent medications, Statistical Methods in Medical Research, (2019) 28(12):3534-3549. SAGE
11. G. Wang*. Review 1: ''Antibiotic Prescribing in Remote Versus Face-to-Face Consultations for Acute Respiratory Infections in English Primary Care: An Observational Study Using TMLE.'' Rapid Reviews Infectious Diseases. (2023) RR
12. A. Jaman, G. Wang, A, Ertefaie, M. Bally, R. Lévesque, RW. Platt, and ME. Schnitzer*. Penalized G-estimation for effect modifier selection in the structural nested mean models for repeated outcomes, in revision, Biometrics, arXiv
13. ME. Schnitzer*, A. Ertefaie, D. Talbot, G. Wang, D. Berger, J. O'Loughlin, M.P. Sylvestre. Longitudinal outcome-adaptive and marginal fused Lasso for confounder selection and model pooling with time-varying treatments
14. G. Wang*, ME. Schnitzer, T. Chen, R. Wang, RW. Platt. A general framework for formulating structured variable selection, Transactions of Machine Learning Research, ISSN: 2835-8856 (2024); OpenReview
15. G. Wang*, ME. Schnitzer, RW. Platt, R. Wang, M. Doris, S. Perreault, Integrating complex selection rules into the latent overlapping group Lasso for constructing coherent prediction models, under review, Statistical in Medicine, arXiv
16. G. Wang*', Y. Lian', A. Yang, RW. Platt, R. Wang, M. Dorias, S. Perreault, M. Schnitzer. Structured learning in Cox models with time-dependent covariates, Statistics in Medicine, 2024; 1-20. doi: 10.1002/sim.10116, Wiley, R package
17. A. Bouchard, F. Bourdeau, J. Roger, VT. Taillefer, N. Sheehan, ME. Schnitzer, G. Wang, IJ. Jean Batiste, and R. Therrien*. Predictive Factors of Detectable Viral Load in HIV Infected Patients, AIDS Research and Human Retroviruses, (2022) 38(7):552-560, Libertpub
18. X. Di, Y. Chi, L. Xiang, G. Wang, B. Liao*.Association between Sitting Time and Urinary Incontinence in the US Population: data from the National Health and Nutrition Examination Survey (NHANES) 2007 to 2018, Heliyon (2024) 10(6):E27764, Elsevier
19. G. Wang, TE. Liao, D. Furfaro, LA. Celi*, KS. Ma*, Extending inference from randomized clinical trials to target populations: a scoping review of transportability methods, arXiv, under review, Journal of the American Medical Informatics Association
20. G. Wang, PJ. Heagerty, IJ. Dahabreh*, Effect score analyses in randomized clinical trials, Journal of American Medical Association. 2024;331(14):1225–1226. doi:10.1001/jama.2024.3376, JAMA
21. G. Wang'*, R. Karlsson'*, J. Krijthe, IJ. Dahabreh, Robust integration of external control data in randomized trials, under review, Biometrics , arXiv
About structured variable selection:
We encourage researchers to thoroughly understand the data before applying any analytical methods. Integrating such information can significantly enhance the performance of the model in use.
For example, when it comes to variable selection, incorporating specific selection rules can lead to improvements in prediction accuracy, a reduced false alarm rate, and, notably, enhanced interpretability of the selected model.
Examples of simple rules can be "if the interaction is chosen, then all or at least one of the main terms must also be selected", "if the subtopic is selected, then the overarching topic should also be chosen", "select at least one gene from each pathway", and "collectively select dummy variables representing a categorical variable."
We have developed unified methods for systematically integrating all available information into the analysis process.
We encourage researchers to thoroughly understand the data before applying any analytical methods. Integrating such information can significantly enhance the performance of the model in use.
For example, when it comes to variable selection, incorporating specific selection rules can lead to improvements in prediction accuracy, a reduced false alarm rate, and, notably, enhanced interpretability of the selected model.
Examples of simple rules can be "if the interaction is chosen, then all or at least one of the main terms must also be selected", "if the subtopic is selected, then the overarching topic should also be chosen", "select at least one gene from each pathway", and "collectively select dummy variables representing a categorical variable."
We have developed unified methods for systematically integrating all available information into the analysis process.